import xarray as xr
import numpy as np
import matplotlib.pyplot as plt
import matplotlib as mplWorking with data that crosses the antimeridian
History | Updated Sep 2023
Background
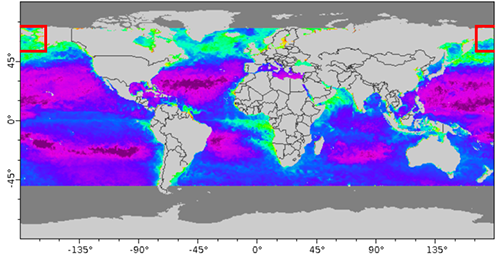
Many datasets use a system where longitude is numbered from -180 to +180 degrees east (see example below). This numbering system presents a problem for researchers working in a region that spans the antimeridian, because the parts of the data end up on the opposite ends of the map.
Objectives
This tutorial will demonstrate how to use datasets with -180 to +180 longitude values to work within regions that cross the antimeridian.
The tutorial demonstrates the following techniques
- Downloading data that crosses the antimeridian from a dataset with -180 to +180 longitude values
- Convert the data to a 0-360 longitude values
- Reordering the longitude axis so that the longitude values are in ascending order
- Visualizing data on a map
Datasets used
NOAA Chlorophyll Gap-filled, Blended NOAA-20 and S-NPP VIIRS, Science Quality, Global, 9km, 2018- recent, Daily
This NOAA dataset blends chlorophyll data from the Visible and Infrared Imager/Radiometer Suite (VIIRS) sensors aboard the Suomi-NPP and NOAA-20 spacecraft. The gaps in the data are then filled using an empirical orthogonal function (DINEOF). The dataset is available from the CoastWatch Central ERDDAP: https://coastwatch.noaa.gov/erddap/griddap/noaacwNPPN20VIIRSSCIDINEOFDaily
Import packages
Define the area to extract
We will extract data for an area in the Bering Sea between Russia and the United States at 176°E to -152°E longitude and 50°N to 70°N latitude.
Set up variables
- Date to extract
- Minimum and maximum values for the longitude and latitude ranges.
my_date = '2023-08-18'
lon_min = -152.
lon_max = 176.
lat_min = 50.
lat_max = 70.Get the SeaWiFS data
Open an xarray dataset object
#url = 'https://coastwatch.noaa.gov/erddap/griddap/noaacwNPPN20VIIRSSCIDINEOFDaily'
url = '/'.join(['https://coastwatch.noaa.gov',
'erddap',
'griddap',
'noaacwNPPN20VIIRSSCIDINEOFDaily'
])
ds = xr.open_dataset(url)
ds<xarray.Dataset>
Dimensions: (time: 1874, altitude: 1, latitude: 2160, longitude: 4320)
Coordinates:
* time (time) datetime64[ns] 2018-05-30T12:00:00 ... 2023-08-24T12:00:00
* altitude (altitude) float64 0.0
* latitude (latitude) float32 89.96 89.88 89.79 ... -89.79 -89.88 -89.96
* longitude (longitude) float32 -180.0 -179.9 -179.8 ... 179.8 179.9 180.0
Data variables:
chlor_a (time, altitude, latitude, longitude) float32 ...
Attributes: (12/77)
_lastModified: 2023-09-04T21:15:13.000Z
_NCProperties: version=2,netcdf=4.7.3,hdf5=1.12.0,
cdm_data_type: Grid
Conventions: CF-1.6, COARDS, ACDD-1.3
creator_email: coastwatch.info@noaa.gov
creator_name: NOAA CoastWatch
... ...
testOutOfDate: now-4days
time_coverage_end: 2023-08-24T12:00:00Z
time_coverage_start: 2018-05-30T12:00:00Z
title: Chlorophyll (Gap-filled DINEOF), NOAA S...
Westernmost_Easting: -179.9583
westernmost_longitude: -180.0Subset the data
We will do this in two steps to make the process easier to follow. 1. Subset the data for date and latitude range 2. Subset the area around the antimeridian * Request data < the limit (-152) on the US side of the antimeridian, i.e. -180 to -152 * Request data > the limit (176) on the Russian side of the antimeridian, i.e. 176 to 180
# The lat1 and lat2 values are used in the slice function
# At first we set them as if the dataset is compliant with standards
lat1 = lat_min
lat2 = lat_max
# Then switch them if latitude values are descending
if ds.latitude[0].item() > ds.latitude[-1].item():
lat1 = lat_max
lat2 = lat_min
# Subset the data in two steps to make it easier to understand
# 1. Subset the date and latitude range
ds_subset = ds['chlor_a'].sel(time=my_date,
method='nearest').sel(latitude=slice(lat1, lat2))
# 2. Subset the around the antimeridian
ds_subset = ds_subset.sel(longitude=(ds.longitude < lon_min)
| (ds.longitude > lon_max)
)
ds_subset<xarray.DataArray 'chlor_a' (altitude: 1, latitude: 240, longitude: 384)>
[92160 values with dtype=float32]
Coordinates:
time datetime64[ns] 2023-08-18T12:00:00
* altitude (altitude) float64 0.0
* latitude (latitude) float32 69.96 69.88 69.79 69.71 ... 50.21 50.12 50.04
* longitude (longitude) float32 -180.0 -179.9 -179.8 ... 179.8 179.9 180.0
Attributes: (12/13)
C_format: %.4g
cell_methods: time:mean(interval:1 day)
colorBarMaximum: 30.0
colorBarMinimum: 0.03
colorBarScale: Log
coverage_content_type: physicalMeasurement
... ...
ioos_category: Ocean Color
long_name: Chlorophyll Concentration, DINEOF Gap-Filled
standard_name: mass_concentration_of_chlorophyll_a_in_sea_water
units: mg m^-3
valid_max: 100.0
valid_min: 0.001Plot the downloaded data
# Make plot area
fig, ax = plt.subplots(figsize=(7, 5))
# Set the color palette
cmap = mpl.cm.get_cmap("jet").copy()
cmap.set_bad(color='gray')
# Show image
shw = ax.imshow(np.log10(ds_subset.squeeze()), cmap=cmap, vmin=-1, vmax=2)
bar = plt.colorbar(shw, shrink=0.65)
# Show plot with labels
bar.set_label('Chlorophyll (log mg m-3)')
plt.show()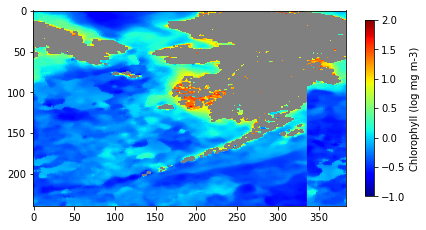
The plot of the data shows a discontinuity
The plot of subsetted data (above) shows that we downloaded the data we requested, but there is a discontinuity on the right side of the map. The data from the Russian side (west of the antimeridian) is mapped to the east of the data on the US side.
To fix the discontinuity, we need to:
* Change the longitude values on the US side of the antimeridian (-180 to -152) to values on the 0-360 longitude indexing system (180-208). * Rearrange the longitude values so that the data on the Russian side is moved to the west of the data on the US side.
Change to 0-360 longitude numbering
ds_360 = ds_subset.assign_coords(longitude=(ds_subset.longitude % 360))
print('minimum lon value =', ds_360.longitude.min().item())
print('minimum lon value =', ds_360.longitude.max().item(), end='\n\n')
print('first value in lon array =', ds_360.longitude[0].item())
print('last value in lon array =', ds_360.longitude[-1].item())minimum lat value = 176.0416717529297
minimum lat value = 207.9583282470703
first value in lat array = 180.0416717529297
last value in lat array = 179.95834350585938Reorder the longitude axis
The output from the cell above shows that the longitude values have been converted to 0-360. However, the lowest longitude value is not at the beginning of the array and the highest longitude value is not at the end of the array.
To rearrange the longitude values, use the roll function of xarray. The roll function will push values along an axis by the number of steps you enter. The values that are “pushed off” of the end of the array will be put at the beginning of the array.
- First we need to find the position where the longitude discontinuity happens, i.e. where the most easterly longitude (208.0) abruptly meets the most easterly longitude value (176.0)
- Next, use the discontinuity position to determine how many positions to roll the longitude array to the right. Apply the number to the
rollfunction.
# This code finds the index where the absolute value between each longitude value
# and the largest longitude value is maximal
discont_index = max(range(len(ds_360.longitude)),
key=lambda i: abs(ds_360.longitude[i] -
ds_360.longitude.max())
)
print('the index marking the discontinuity is:', discont_index, end='\n\n')
# Substract the discontinuity position from the length of the array
# to obtain the number of positions to roll the longitude axis
postions_to_roll = len(ds_360.longitude) - discont_index
# Roll the dataset
ds_rolled = ds_360.roll(longitude=postions_to_roll, roll_coords=True)
print('minimum lon value =', ds_rolled.longitude.min().item())
print('minimum lon value =', ds_rolled.longitude.max().item(), end='\n\n')
print('first value in lon array =', ds_rolled.longitude[0].item())
print('last value in lon array =', ds_rolled.longitude[-1].item())the index marking the discontinuity is: 336
minimum lon value = 176.0416717529297
minimum lon value = 207.9583282470703
first value in lon array = 176.0416717529297
last value in lon array = 207.9583282470703The output from the cell above shows that the longitude values have been converted to 0-360, and that the lowest longitude value is at the beginning of the array and the highest longitude value is at the end of the array.
Plot the data
The discontinuity has been corrected!
# Make plot area
fig, ax = plt.subplots(figsize=(7, 5))
# Set the color palette
cmap = mpl.cm.get_cmap("jet").copy()
cmap.set_bad(color='gray')
# show image
shw = ax.imshow(np.log10(ds_rolled.squeeze()),
cmap=cmap,
vmin=-1,
vmax=2)
bar = plt.colorbar(shw, shrink=0.65)
# show plot with labels
bar.set_label('Chlorophyll (log mg m-3)')
plt.show()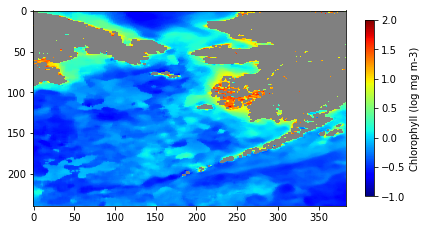
Save the corrected dataset as a netCDF file
ds_rolled.to_netcdf('data_corrected_0_to_360.nc')