from IPython.display import clear_output
import pandas as pd
import numpy as np
import os
import warnings
import cartopy.crs as ccrs
import cartopy.feature as cfeature
from cartopy.mpl.ticker import LongitudeFormatter, LatitudeFormatter
import matplotlib.pyplot as plt
import xarray as xr
warnings.filterwarnings('ignore')Matchup satellite data to ship, glider, or animal tracks
Overview
History | Updated July 2024
In this exercise you will extract satellite data around a set of points defined by longitude, latitude, and time coordinates, like that produced by an animal telemetry tag, and ship track, or a glider tract.
Please note that there may be more efficient ways, more Pythonic ways, to accomplish the tasks in this tutorial. The tutorial was developed to be easier to follow for less experienced users of Python.
The exercise demonstrates the following techniques:
- Loading data from a tab- or comma-separated file
- Plotting the latitude/longitude points onto a map
- Extracting satellite data along a track
- Saving results as a CSV file
- Plotting the satellite data onto a map
Datasets used:
- Chlorophyll a concentration from the European Space Agency’s Ocean Colour Climate Change Initiative Monthly dataset v6.0
- Yellowfin tuna telemetry track data that was developed as part of the Palmyra Bluewater Research (PBR) project (https://portal.atn.ioos.us/?ls=861Wqpd2#metadata/1f877c4c-7b50-49f5-be86-3354664e0cff/project). This example track used in the tutorial is from May 2022 to November 2022 and accessed via the Animal Telemetry Network (ATN) data portal.
Python packages used:
- pandas (reading and analyzing data)
- numpy (data analysis, manipulation)
- xarray (multi-dimensional data analysis, manipulation)
- matplotlib (mapping)
- cartopy (mapping)
- datetime (date manipulation)
Import the required Python modules
Download animal track data from the ATN website
The data you need for this exercise are already in the GitHub repository
The following step are show you how to download the data yourself
ATN Portal
- Follow the link to the ATN website: https://portal.atn.ioos.us/?ls=O6vlufm7#map
- On the right navigational panel, look for the “Palmyra Bluewater Research (PBR) Megafauna Movement Ecology Project, 2022-2023” tab.
- Within the tab, scroll to find the telemetry tag labeled “Yellowfin tuna (233568)”.
- Click the search icon (maginifying glass) label next to the label to zoom into the area the area of the animal track.
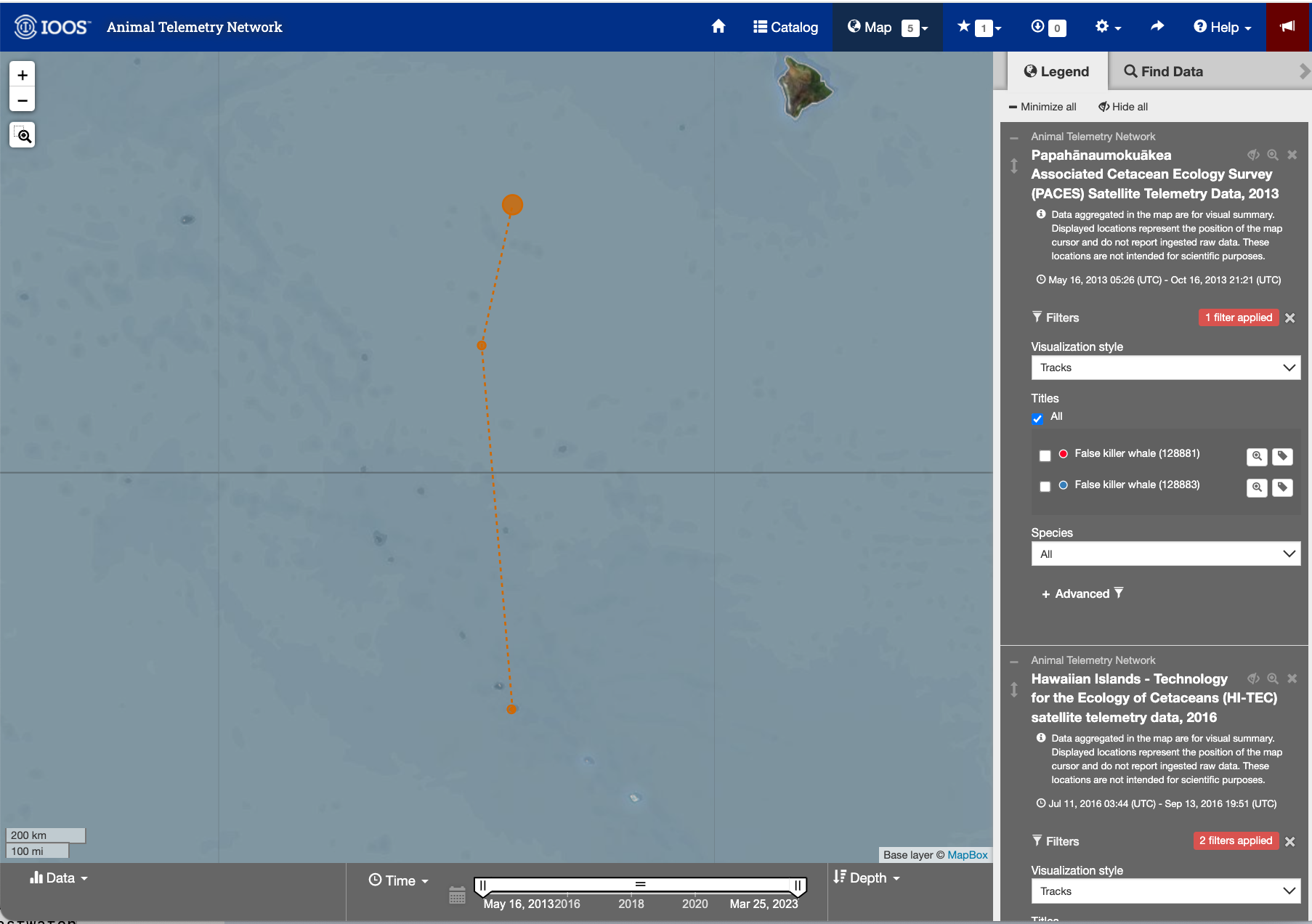
Image of the ATN portal webpage
Detail page for the Yellowfin tuna (233568)
- Next click the tag icon to the right of the search icon.
- This page shows you details about the animal and the track.
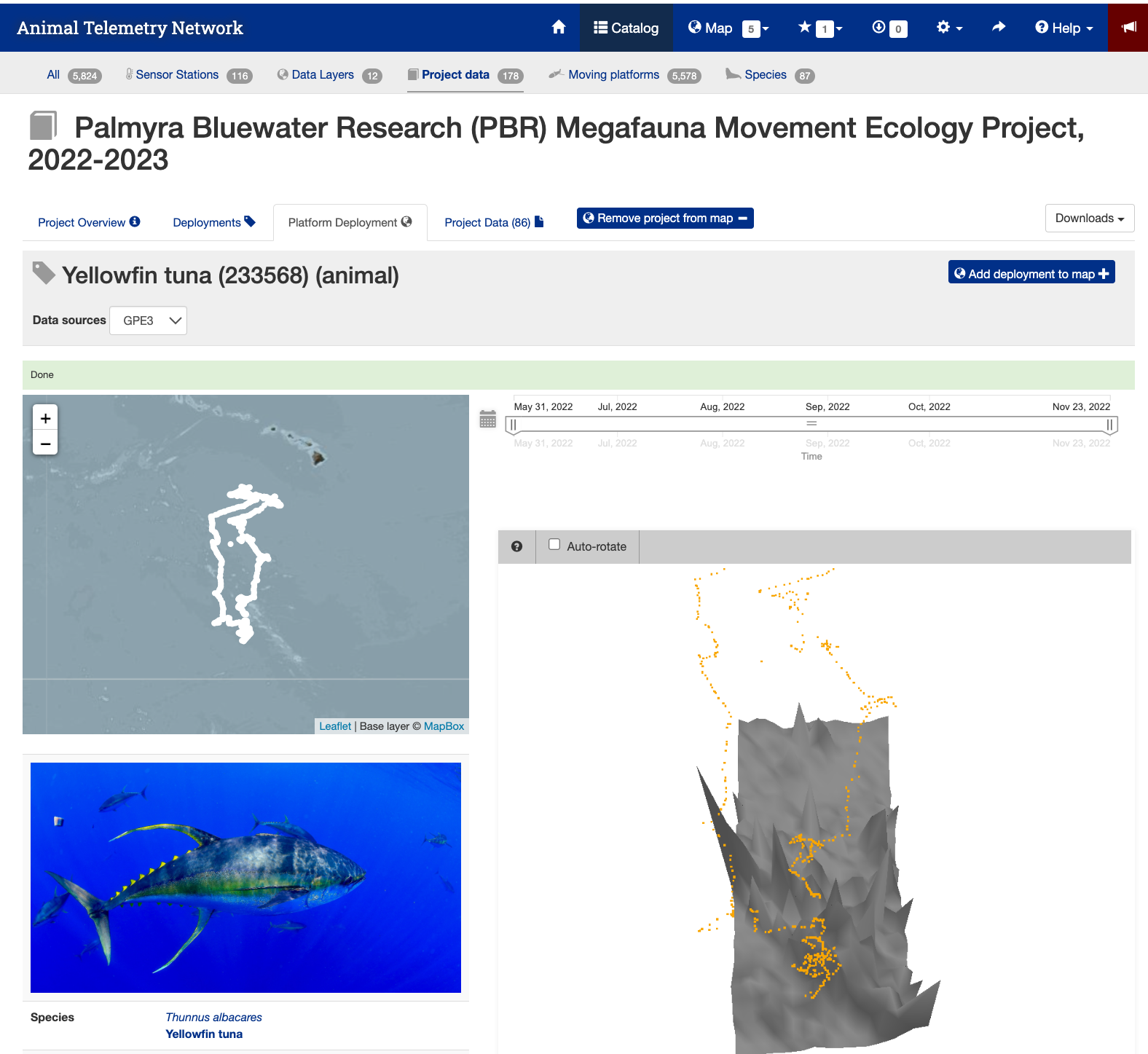
Image of the detail page for the Yellowfin Tuna #233568
Data Download Page
- Press the “Project Data” tab near the top of the webpage to bring up the data file list.
- Search for the animal id number (233568).
- Click on the link “THUALB_2022_04-PTT_233568.zip (8.6 MB)” to download the data.
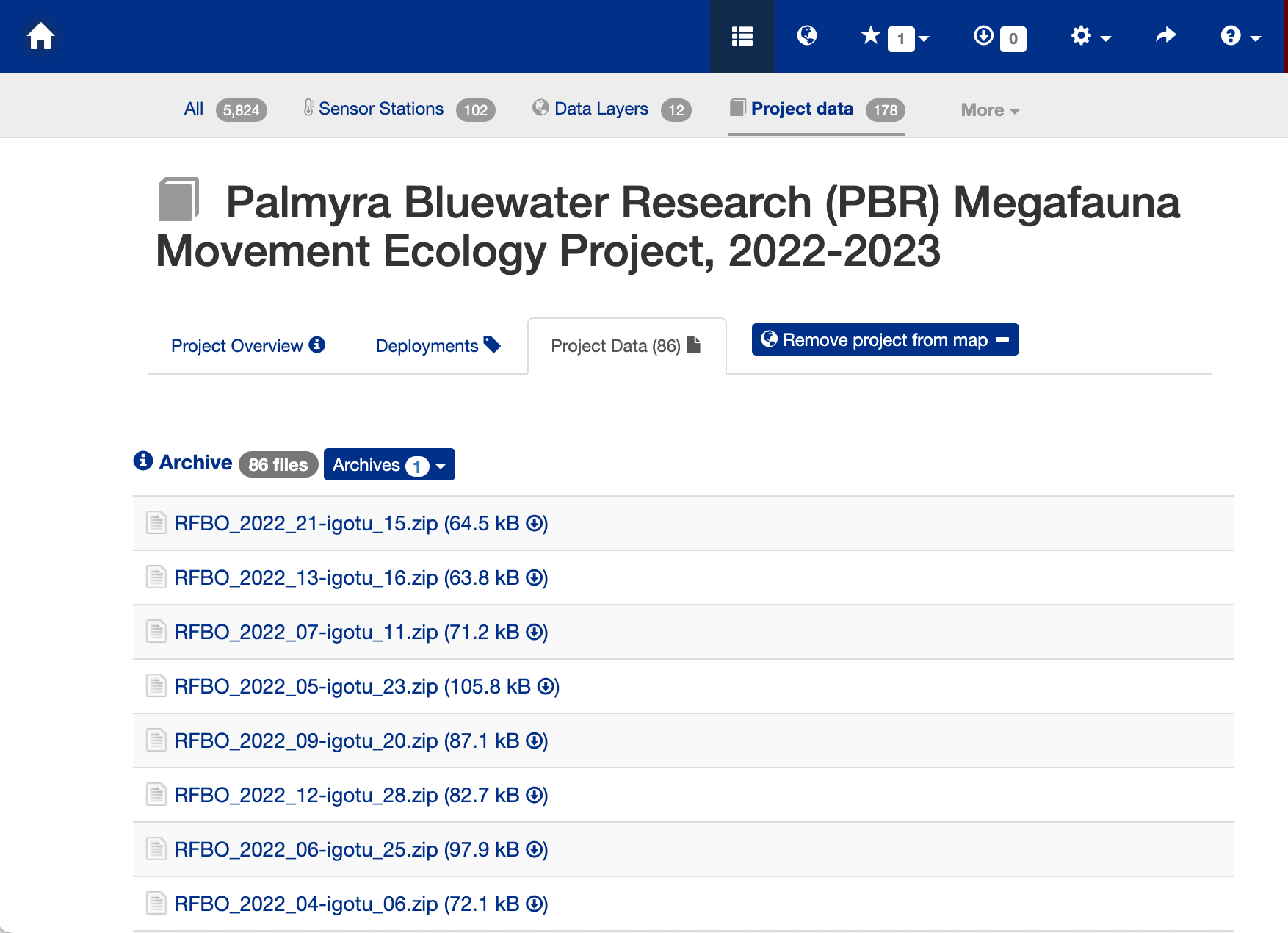
Data download page
The Downloaded Data File
- The download will be a zip file. You will need to unzip it.
- The unzipped file folder contains many ancillary data files, but the one you are looking for is the CSV file (THUALB_2022_04-233568-4-GPE3.csv).
- You don’t have to move this file into the data folder for this exercise. It is already there.
- But if you download the file for a different animal track you would need to put the CSV file into the folder.
Load the track data into a Pandas data frame
Below, the track data will load using the Pandas “read_csv” method. * Then use the “.head()” method to view the column names and the first few rows of data.
track_path = os.path.join('..',
'data',
'THUALB_2022_04-233568-5-GPE3.csv')
track_df = pd.read_csv(track_path, skiprows=4)
track_df.head(2)| DeployID | Ptt | Date | Most Likely Latitude | Most Likely Longitude | Observation Type | Observed SST | Satellite SST | Observed Depth | Bathymetry Depth | Observation LL (MSS) | Observation Score | Sunrise | Sunset | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | THUALB_2022_04 | 233568 | 31-May-2022 19:00:00 | 5.875 | -162.125 | User | NaN | NaN | NaN | NaN | NaN | 68.605853 | NaN | NaN |
| 1 | THUALB_2022_04 | 233568 | 01-Jun-2022 00:00:00 | 5.875 | -162.100 | None | NaN | NaN | NaN | NaN | NaN | NaN | 01-Jun-2022 16:33:04 | 02-Jun-2022 04:59:36 |
## Convert the date strings into the Python datetime date objects * The dates are loaded from the CSV file as strings. * Converting the dates to Python datetime date object gives us flexibility to manipulate the dataframe.
track_df['Date'] = pd.to_datetime(track_df['Date'], format='%d-%b-%Y %H:%M:%S')
track_df.head(2)| DeployID | Ptt | Date | Most Likely Latitude | Most Likely Longitude | Observation Type | Observed SST | Satellite SST | Observed Depth | Bathymetry Depth | Observation LL (MSS) | Observation Score | Sunrise | Sunset | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | THUALB_2022_04 | 233568 | 2022-05-31 19:00:00 | 5.875 | -162.125 | User | NaN | NaN | NaN | NaN | NaN | 68.605853 | NaN | NaN |
| 1 | THUALB_2022_04 | 233568 | 2022-06-01 00:00:00 | 5.875 | -162.100 | None | NaN | NaN | NaN | NaN | NaN | NaN | 01-Jun-2022 16:33:04 | 02-Jun-2022 04:59:36 |
Clean up the dataframe
- Remove the columns with non-numerical data.
- Rename the “Most Likely Latitude” and “Most Likely Longitude” columns to make the easier to work with.
- The track data has longitude values into -180/+180 format. However, the satellite dataset we are using has longitude values into 0/360 format. So, convert the track data to 0/360 format so that the longitude formats are consistent.
del track_df['DeployID']
del track_df['Ptt']
del track_df['Sunrise']
del track_df['Sunset']
del track_df['Observation Type']
track_df.rename(columns={"Most Likely Latitude": "Latitude",
"Most Likely Longitude": "Longitude"}, inplace=True)
# Convert lontitudes to 0~360 (Re-center map to the dateline)
track_df.Longitude = track_df.Longitude + 360
track_df.head(2)| Date | Latitude | Longitude | Observed SST | Satellite SST | Observed Depth | Bathymetry Depth | Observation LL (MSS) | Observation Score | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 2022-05-31 19:00:00 | 5.875 | 197.875 | NaN | NaN | NaN | NaN | NaN | 68.605853 |
| 1 | 2022-06-01 00:00:00 | 5.875 | 197.900 | NaN | NaN | NaN | NaN | NaN | NaN |
Bin multiple observations from each day into daily mean values
The track data has multiple longitude/latitude/time points for each date. That temporal resolution is much higher than the daily and month satellite datasets that are available. So, let’s reduce the multiple daily values for the animal track data to a single value for each day. The code below creates a new dataframe that bins data for each date and calculates the mean for selected columns. * This process will move the “Date” column to the index column (most left column)
df = track_df.resample('D', on='Date').mean()
df.head()| Latitude | Longitude | Observed SST | Satellite SST | Observed Depth | Bathymetry Depth | Observation LL (MSS) | Observation Score | |
|---|---|---|---|---|---|---|---|---|
| Date | ||||||||
| 2022-05-31 | 5.87500 | 197.8750 | NaN | NaN | NaN | NaN | NaN | 68.605853 |
| 2022-06-01 | 5.88500 | 198.0000 | 28.2 | 28.324583 | 85.666667 | 2908.0 | NaN | 58.239984 |
| 2022-06-02 | 5.92500 | 197.9500 | 28.3 | 28.305416 | 64.666667 | 3778.0 | NaN | 93.446222 |
| 2022-06-03 | 5.92500 | 197.8550 | 28.1 | 28.310833 | 53.666667 | 3778.0 | NaN | 71.484871 |
| 2022-06-04 | 5.98125 | 197.8125 | 28.2 | 28.320000 | 43.000000 | 3778.0 | NaN | 82.962634 |
Plot the track on a map
plt.figure(figsize=(14, 10))
# Label axes of a Plate Carree projection with a central longitude of 180:
ax1 = plt.subplot(211, projection=ccrs.PlateCarree(central_longitude=180))
# Use the lon and lat ranges to set the extent of the map
# the 120, 260 lon range will show the whole Pacific
# the 15, 55 lat range with capture the range of the data
ax1.set_extent([180, 221, 0, 26], ccrs.PlateCarree())
# Set the tick marks to be slightly inside the map extents
ax1.set_xticks(range(180, 221, 10), crs=ccrs.PlateCarree())
ax1.set_yticks(range(0, 26, 10), crs=ccrs.PlateCarree())
# Add feature to the map
ax1.add_feature(cfeature.LAND, facecolor='0.6')
ax1.coastlines()
# Format the lat and lon axis labels
lon_formatter = LongitudeFormatter(zero_direction_label=True)
lat_formatter = LatitudeFormatter()
ax1.xaxis.set_major_formatter(lon_formatter)
ax1.yaxis.set_major_formatter(lat_formatter)
# Bring the lon and lat data into a numpy array
x, y = df.Longitude.to_numpy(), df.Latitude.to_numpy()
ax1 = plt.plot(x, y, transform=ccrs.PlateCarree(), color='blue')
# start point in green star
ax1 = plt.plot(x[0], y[0],
marker='*',
color='g',
label='start',
transform=ccrs.PlateCarree(),
markersize=10)
# end point in red X
ax1 = plt.plot(x[-1], y[-1],
marker='X',
color='r',
label='end',
transform=ccrs.PlateCarree(),
markersize=10)
plt.legend()
plt.title('Yellowfin Tuna Track (PTT 233568)', fontsize=20)
plt.show()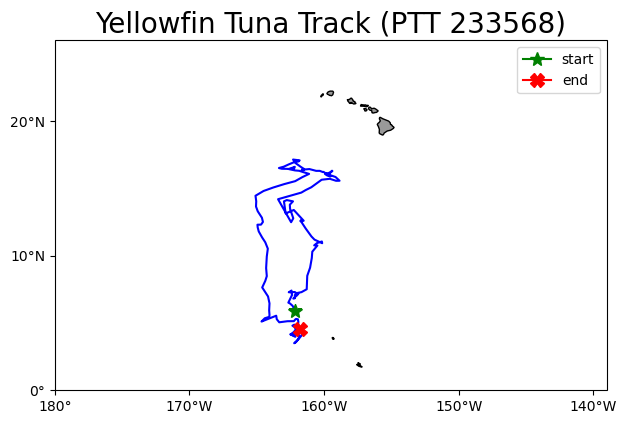
Extract data from a satellite dataset corresponding to points on the track
We are going to download data from an ERDDAP server using the following steps: * Select a dataset on an ERDDAP server * Open the dataset using the Xarray module * Loop though the track data and pull out the date, latitude and longitude coordinates from each row * Insert these coordinates into the Xarray open-dataset object to subset and download the satellite data that corresponds to the coordinates. * Add the satellite data to the track dataset.
Select a dataset
We’ll use the European Space Agency’s OC-CCI product (https://climate.esa.int/en/projects/ocean-colour/) to obtain chlorophyll data. This is a merged product that blends data from many ocean color sensors to create a long time series (1997-present) with better spatial coverage than any single sensor.
Ideally we would use a daily dataset, selecting the day that corresponds to the track data date. However, chlorophyll measurements can have a lot of missing data, primarily due to cloud cover. To reduce data gaps and improve the likelihood of data for our matchups, we can use a dataset that combines all of the data from each month into the monthly average.
The ERDDAP URL to the monthly version of the OC-CCI product is below:
https://oceanwatch.pifsc.noaa.gov/erddap/griddap/esa-cci-chla-monthly-v6-0
Open the satellite data in Xarray
- Use the ERDDAP URL with no extension (e.g. without .html or .graph…). This is the OPeNDAP URL, which allows viewing the dataset metadata and, when you select the data you want, downloading the data.
- Use the Xarray “open_dataset” function then view the metadata
erddap_url = '/'.join(['https://oceanwatch.pifsc.noaa.gov',
'erddap',
'griddap',
'esa-cci-chla-monthly-v6-0'])
ds = xr.open_dataset(erddap_url)
ds<xarray.Dataset>
Dimensions: (time: 316, latitude: 4320, longitude: 8640)
Coordinates:
* time (time) datetime64[ns] 1997-09-04 ... 2023-12-01
* latitude (latitude) float64 89.98 89.94 89.9 ... -89.94 -89.98
* longitude (longitude) float64 0.02083 0.0625 ... 359.9 360.0
Data variables:
chlor_a (time, latitude, longitude) float32 ...
MERIS_nobs_sum (time, latitude, longitude) float32 ...
MODISA_nobs_sum (time, latitude, longitude) float32 ...
OLCI_A_nobs_sum (time, latitude, longitude) float32 ...
OLCI_B_nobs_sum (time, latitude, longitude) float32 ...
SeaWiFS_nobs_sum (time, latitude, longitude) float32 ...
VIIRS_nobs_sum (time, latitude, longitude) float32 ...
chlor_a_log10_bias (time, latitude, longitude) float32 ...
chlor_a_log10_rmsd (time, latitude, longitude) float32 ...
total_nobs_sum (time, latitude, longitude) float32 ...
Attributes: (12/53)
cdm_data_type: Grid
comment: See summary attribute
Conventions: CF-1.7, COARDS, ACDD-1.3
creation_date: Thu Jan 18 09:04:18 2024
creator_email: help@esa-oceancolour-cci.org
creator_name: Plymouth Marine Laboratory
... ...
time_coverage_end: 2023-12-01T00:00:00Z
time_coverage_resolution: P1M
time_coverage_start: 1997-09-04T00:00:00Z
title: Chlorophyll a concentration, ESA OC CC...
tracking_id: abd52a4c-7009-464f-b1eb-958f7d333a1d
Westernmost_Easting: 0.020833333333314386Opening the dataset in Xarray lets you look at the dataset metadata.
* The metadata are listed above. * No data is downloaded until you request it.
From the metadata you can view: * The coordinates (time, latitude and longitude) that you will use to select the data to download. * A list of ten data variables. For this exercise, we want the “chlor_a” variable. If you want, you can find out about each variable with clicking the page icon to the right of each variable name.
A note on dataset selection
We have preselected the OC-CCI monthly dataset because we know it will work with this exercise. If you were selecting datasets on your own, you would want to check out the dataset to determine if its spatial and temporal coverages are suitable for your application.
You can find that information above by clicking the right arrow next to “Attribute”. Then look through the list to find: * ‘time_coverage_start’ and ‘time_coverage_end’: the time range * ‘geospatial_lat_min’ and ‘geospatial_lat_max’: the latitude range * ‘geospatial_lon_min’ and ‘geospatial_lon_max’: the longitude range
There are a lot of metadata attributes to look through. We can make it easier with a little code to print out the metadata of interest. Then compare these ranges to those found in your track data.
print('Temporal and spatial ranges of the satellite dataset')
print('time range', ds.attrs['time_coverage_start'],
ds.attrs['time_coverage_end'])
print('latitude range', ds.attrs['geospatial_lat_min'],
ds.attrs['geospatial_lat_max'])
print('longitude range', ds.attrs['geospatial_lon_min'],
ds.attrs['geospatial_lon_max'])
print(' ')
print('Temporal and spatial ranges of the track data')
print('time range', df.index.min(), df.index.max())
print('latitude range',
round(df.Latitude.min(), 2), round(df.Latitude.max(), 2))
print('longitude range',
round(df.Longitude.min(), 2), round(df.Longitude.max(), 2))Temporal and spatial ranges of the satellite dataset
time range 1997-09-04T00:00:00Z 2023-12-01T00:00:00Z
latitude range -89.97916666666666 89.97916666666667
longitude range 0.020833333333314386 359.97916666666663
Temporal and spatial ranges of the track data
time range 2022-05-31 00:00:00 2022-11-23 00:00:00
latitude range 3.5 17.14
longitude range 194.91 201.15Download the satellite data that corresponds to each track location
- First, create new columns in the df dataframe to hold the downloaded data. Fill each row with nan.
- Download the chlorophyll data, time, latitude, and longitude from the ERDDAP satellite dataset.
- Consolidate data adding the downloaded satellite data to the track data (df) dataframe.
# Add new columns to the dataframe
df[["erddap_date", "matched_lat", "matched_lon", "matched_chla"]] = np.nan
# Subset the satellite data
for i in range(0, len(df)):
clear_output(wait=True)
print(i+1, 'of', len(df))
# Download the satellite data and put into a temperary dataframe
temp_ds = ds['chlor_a'].sel(time='{0:%Y-%m-%d}'.format(df.index[1]),
latitude=df.loc[df.index[i], 'Latitude'],
longitude=df.loc[df.index[i], 'Longitude'],
method='nearest'
)
# Consolidate the data
df.loc[df.index[i], ["erddap_date", "matched_lat",
"matched_lon", "matched_chla"]
] = [temp_ds.time.values,
np.round(temp_ds.latitude.values, 5), # round 5 dec
np.round(temp_ds.longitude.values, 5), # round 5 dec
np.round(temp_ds.values, 2) # round 2 decimals
]
print(df.loc[df.index[i], ["erddap_date", "matched_lat",
"matched_lon", "matched_chla"]])
177 of 177
erddap_date 2022-06-01T00:00:00.000000000
matched_lat 4.52083
matched_lon 198.22917
matched_chla 0.22
Name: 2022-11-23 00:00:00, dtype: objectSave your work
- Since we moved the Date column to the index, be sure to use index=True
df.to_csv('chl_matchup_tuna233568.csv', index=True, encoding='utf-8')Plot chlorophyll matchup data onto a map
First plot a histogram of the chlorophyll data
print('Range:', df.matched_chla.min(), df.matched_chla.max())
_ = df.matched_chla.hist(bins=40)Range: 0.03999999910593033 0.4699999988079071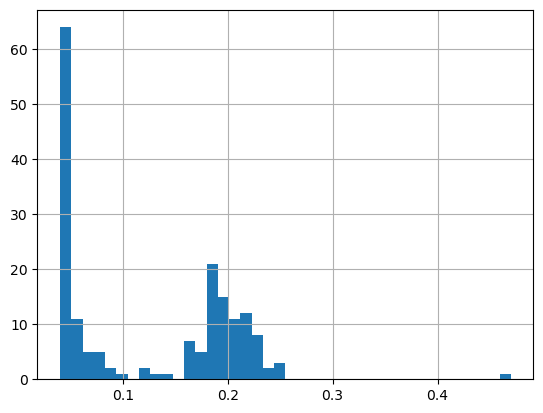
The range of chlorophyll values can be large, with lots of very low values and a few very high values. Using a linear color bar, most of the lower values would have the same color. * To better visualize the data, we often plot the log or log10 of chlorophyll.
Plot a histogram of the log of the chlorophyll data
print('Range:',
np.log(df.matched_chla.min()),
np.log(df.matched_chla.max()))
_ = np.log(df.matched_chla).hist(bins=40)Range: -3.218875847219943 -0.7550225868144006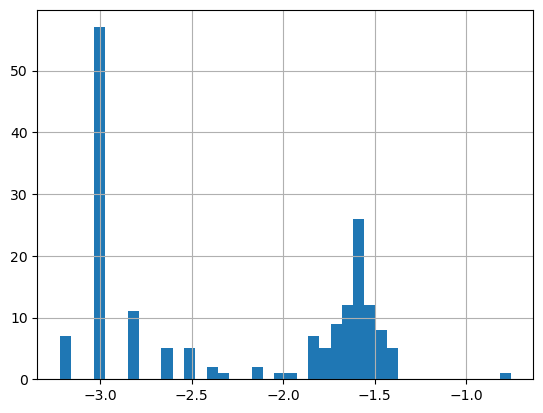
- The logarithmic transformation displays the range of values across the color bar range (above).
Map the chlorophyll data
plt.figure(figsize=(14, 10))
# Label axes of a Plate Carree projection with a central longitude of 180:
# set the projection
ax1 = plt.subplot(211, projection=ccrs.PlateCarree(central_longitude=180))
# Use the lon and lat ranges to set the extent of the map
# the 120, 260 lon range will show the whole Pacific
# the 15, 55 lat range with capture the range of the data
ax1.set_extent([180, 221, 0, 26], ccrs.PlateCarree())
# Set the tick marks to be slightly inside the map extents
ax1.set_xticks(range(180, 221, 10), crs=ccrs.PlateCarree())
ax1.set_yticks(range(0, 26, 10), crs=ccrs.PlateCarree())
# Add geographical features
ax1.add_feature(cfeature.LAND, facecolor='0.6')
ax1.coastlines()
# format the lat and lon axis labels
lon_formatter = LongitudeFormatter(zero_direction_label=True)
lat_formatter = LatitudeFormatter()
ax1.xaxis.set_major_formatter(lon_formatter)
ax1.yaxis.set_major_formatter(lat_formatter)
# build and plot coordinates onto map
x,y = list(df.Longitude), list(df.Latitude)
ax1 = plt.scatter(x, y, transform=ccrs.PlateCarree(),
marker='o',
c=np.log(df.matched_chla),
cmap=plt.get_cmap('jet')
)
ax1=plt.plot(x[0],y[0],marker='*', label='start',transform=ccrs.PlateCarree(), markersize=10)
ax1=plt.plot(x[-1],y[-1],marker='X', label='end',transform=ccrs.PlateCarree(), markersize=10)
# control color bar values spacing
levs2 = np.arange(-3, -1, 0.5)
cbar=plt.colorbar(ticks=levs2, shrink=0.75, aspect=10)
#cbar=plt.colorbar(shrink=0.75, aspect=10)
cbar.set_label("Chl a (mg/m^3)", size=15, labelpad=20)
# set the labels to be exp(levs2) so the label reflect values of chl-a, not log(chl-a)
cbar.ax.set_yticklabels(np.around(np.exp(levs2), 2), size=10)
plt.legend()
plt.title("Chlorophyll Matchup to Yellowfin Tuna Track (233568)", size=15)
plt.show()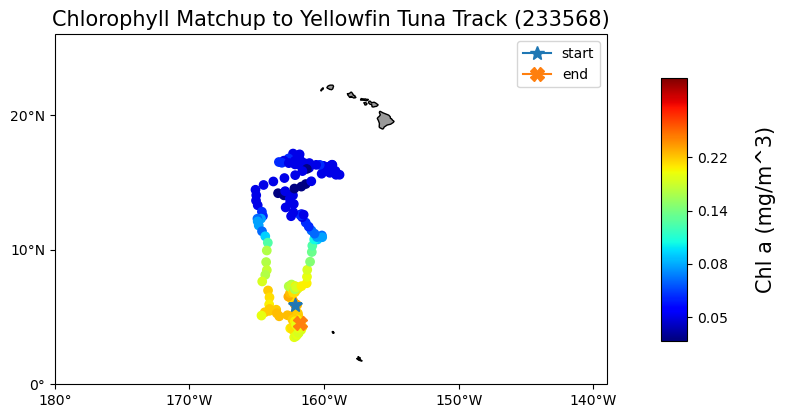
On your own!
Exercise 1:
Repeat the steps above with a different dataset. For example, extract sea surface temperature data using the NOAA Geo-polar Blended Analysis SST, GHRSST dataset: https://coastwatch.pfeg.noaa.gov/erddap/griddap/nesdisGeoPolarSSTN5NRT_Lon0360.html
* This dataset is a different ERDDAP; It has a different base URL and dataset ID and variable name. * Check the metadata to make sure the dataset covers the spatial and temporal range of the track dataset.
Exercise 2:
Go to an ERDDAP of your choice, find a dataset of interest, generate the URL, copy it and edit the script above to run a match up on that dataset. To find other ERDDAP servers, you can use this search engine: http://erddap.com/
* This dataset will likely be on a different base URL and dataset ID and variable name. * Check the metadata to make sure the dataset covers the spatial and temporal range of the track dataset.
Optional
Repeat the steps above with a daily version of the OC-CCI dataset to see how cloud cover can reduce the data you retrieve. https://oceanwatch.pifsc.noaa.gov/erddap/griddap/CRW_sst_v1_0.html
